In 2013, Mikolov published his paper on word2vec. What’s the main idea of word2vec? It is composed of two algorithms: Skip-grams and CBOW (continuous bag of words). In this new paper – Distributed Representations of Sentences and Documents, he points out “Despite their popularity, bag-of-words features have two major weaknesses: they lose the ordering of the words and they also ignore semantics of the words. For example, “powerful,” “strong”
and “Paris” are equally distant.”, hence he proposed “Paragraph Vector, an unsupervised algorithm that learns fixed-length feature representations from variable-length pieces of texts, such as sentences, paragraphs, and documents.” The new algorithm he/Google developed “represents each document by a dense vector which is trained to predict words in the document.”
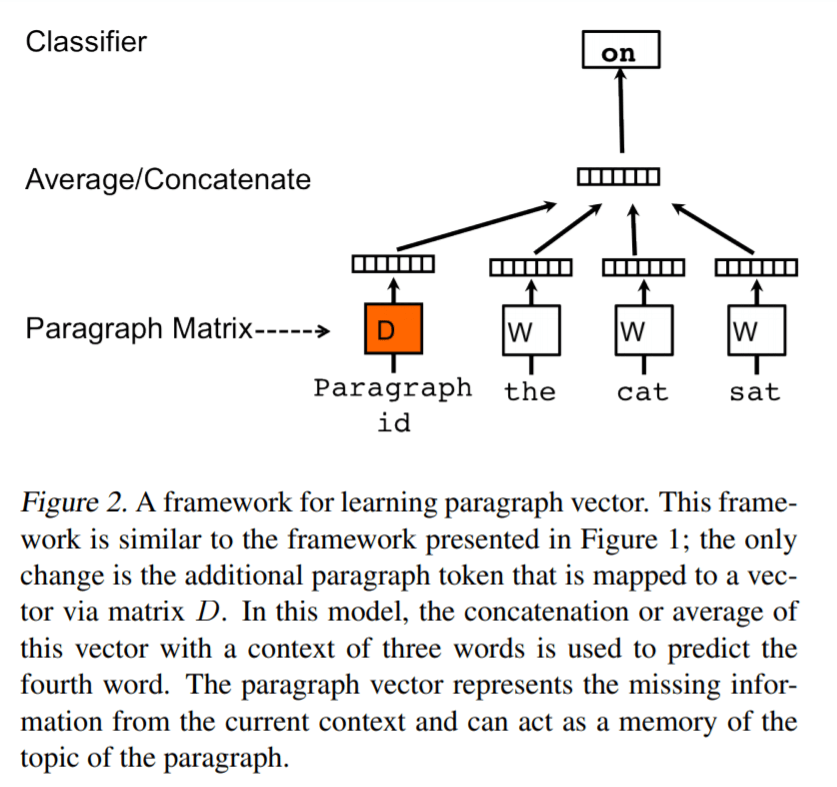
As the author pointed out Doc2Vec is inspired or based on Word2Vec as illustrated below:
Experiments are conducted on IMDB movie review set of data, and the following is the result:

Now we’ll try to replicate the LDA analysis on IMDB movie review data using gensim.
LDA parameters: chunksize controls how man documents are processed at a time, one can set it to be chunksize = 2000; passes controls how often we train the model on the entire corpus. same as epochs. alpha=’auto’ and eta=’auto’.
#replicate makolov lda paper
import collections
SentimentDocument = collections.namedtuple('SentimentDocument', 'words tags split sentiment')
import io
import re
import tarfile
import os.path
import smart_open
import gensim.utils
def download_dataset(url='http://ai.stanford.edu/~amaas/data/sentiment/aclImdb_v1.tar.gz'):
fname = url.split('/')[-1]
if os.path.isfile(fname):
return fname
# Download the file to local storage first.
with smart_open.open(url, "rb", ignore_ext=True) as fin:
with smart_open.open(fname, 'wb', ignore_ext=True) as fout:
while True:
buf = fin.read(io.DEFAULT_BUFFER_SIZE)
if not buf:
break
fout.write(buf)
return fname
def create_sentiment_document(name, text, index):
_, split, sentiment_str, _ = name.split('/')
sentiment = {'pos': 1.0, 'neg': 0.0, 'unsup': None}[sentiment_str]
if sentiment is None:
split = 'extra'
tokens = gensim.utils.to_unicode(text).split()
return SentimentDocument(tokens, [index], split, sentiment)
def extract_documents():
fname = download_dataset()
index = 0
with tarfile.open(fname, mode='r:gz') as tar:
for member in tar.getmembers():
if re.match(r'aclImdb/(train|test)/(pos|neg|unsup)/\d+_\d+.txt$', member.name):
member_bytes = tar.extractfile(member).read()
member_text = member_bytes.decode('utf-8', errors='replace')
assert member_text.count('\n') == 0
yield create_sentiment_document(member.name, member_text, index)
index += 1
alldocs = list(extract_documents())
alldocs[0]
train_docs = [doc for doc in alldocs if doc.split == 'train']
test_docs = [doc for doc in alldocs if doc.split == 'test']
print(f'{len(alldocs)} docs: {len(train_docs)} train-sentiment, {len(test_docs)} test-sentiment')
import multiprocessing
from collections import OrderedDict
import gensim.models.doc2vec
assert gensim.models.doc2vec.FAST_VERSION > -1, "This will be painfully slow otherwise"
from gensim.models.doc2vec import Doc2Vec
common_kwargs = dict(
vector_size=100, epochs=20, min_count=2,
sample=0, workers=multiprocessing.cpu_count(), negative=5, hs=0,
)
simple_models = [
# PV-DBOW plain
Doc2Vec(dm=0, **common_kwargs),
# PV-DM w/ default averaging; a higher starting alpha may improve CBOW/PV-DM modes
Doc2Vec(dm=1, window=10, alpha=0.05, comment='alpha=0.05', **common_kwargs),
# PV-DM w/ concatenation - big, slow, experimental mode
# window=5 (both sides) approximates paper's apparent 10-word total window size
Doc2Vec(dm=1, dm_concat=1, window=5, **common_kwargs),
]
for model in simple_models:
model.build_vocab(alldocs)
print(f"{model} vocabulary scanned & state initialized")
models_by_name = OrderedDict((str(model), model) for model in simple_models)
from gensim.test.test_doc2vec import ConcatenatedDoc2Vec
models_by_name['dbow+dmm'] = ConcatenatedDoc2Vec([simple_models[0], simple_models[1]])
models_by_name['dbow+dmc'] = ConcatenatedDoc2Vec([simple_models[0], simple_models[2]])
#Predicative Evaluation Methods
import numpy as np
import statsmodels.api as sm
from random import sample
def logistic_predictor_from_data(train_targets, train_regressors):
"""Fit a statsmodel logistic predictor on supplied data"""
logit = sm.Logit(train_targets, train_regressors)
predictor = logit.fit(disp=0)
# print(predictor.summary())
return predictor
def error_rate_for_model(test_model, train_set, test_set):
"""Report error rate on test_doc sentiments, using supplied model and train_docs"""
train_targets = [doc.sentiment for doc in train_set]
train_regressors = [test_model.dv[doc.tags[0]] for doc in train_set]
train_regressors = sm.add_constant(train_regressors)
predictor = logistic_predictor_from_data(train_targets, train_regressors)
test_regressors = [test_model.dv[doc.tags[0]] for doc in test_set]
test_regressors = sm.add_constant(test_regressors)
# Predict & evaluate
test_predictions = predictor.predict(test_regressors)
corrects = sum(np.rint(test_predictions) == [doc.sentiment for doc in test_set])
errors = len(test_predictions) - corrects
error_rate = float(errors) / len(test_predictions)
return (error_rate, errors, len(test_predictions), predictor)
# Bulk Training & Per-Model Evaluation, take an hour to train
from collections import defaultdict
error_rates = defaultdict(lambda: 1.0) # To selectively print only best errors achieved
from random import shuffle
shuffled_alldocs = alldocs[:]
shuffle(shuffled_alldocs)
for model in simple_models:
print(f"Training {model}")
model.train(shuffled_alldocs, total_examples=len(shuffled_alldocs), epochs=model.epochs)
print(f"\nEvaluating {model}")
err_rate, err_count, test_count, predictor = error_rate_for_model(model, train_docs, test_docs)
error_rates[str(model)] = err_rate
print("\n%f %s\n" % (err_rate, model))
for model in [models_by_name['dbow+dmm'], models_by_name['dbow+dmc']]:
print(f"\nEvaluating {model}")
err_rate, err_count, test_count, predictor = error_rate_for_model(model, train_docs, test_docs)
error_rates[str(model)] = err_rate
print(f"\n{err_rate} {model}\n")
print("Err_rate Model")
for rate, name in sorted((rate, name) for name, rate in error_rates.items()):
print(f"{rate} {name}")
doc_id = np.random.randint(len(simple_models[0].dv)) # Pick random doc; re-run cell for more examples
print(f'for doc {doc_id}...')
for model in simple_models:
inferred_docvec = model.infer_vector(alldocs[doc_id].words)
print(f'{model}:\n {model.dv.most_similar([inferred_docvec], topn=3)}')
It’s interesting that the gensim engineer claimed that they can’t replicate the 7% error rate in the paper by Makolov, the best is only 10%. However it’s pretty good a result too.
Next step for me is to apply the same on my own custom dataset on hydrogen.